An R reproducibility toolkit for the practical researcher > Day 1 > 03: Reporting with R Markdown
03: Reporting with R Markdown
By Paola Corrales and Elio Campitelli
R Markdown files
An R Markdown file is a plain text file, with some rules and special syntax that allow you to write code and text together. When it is “knitted,” the code will be evaluated and executed and the text formatted so that it creates a reproducible report or document that is nice to read and contains all your work.
This is really critical to reproducibility since it automates the creation of a paper or report, which also has the side effect of saving time. Since the document will recreate the figures and text each time you knit it, you won’t need to copy-paste plots between R and Word, LaTeX or whatever tool you are using to write your report each time you make some trivial change.
Now let’s create a new R Markdown file. In RStudio you can use the menu bar:
File → New File → R Markdown
A new window will open where you can optionally complete with your name, author and the output format. Let try HTML output first. A new file with a template document will open up when you click OK. This includes a brief demonstration of R Markdown capabilities. You can try to “knit” the file to see the output document.
Your turn
-
Create a new R Markdown file. You can choose the output format.
-
Knit the sample document.
-
Compare the “source file” with the “output file”. Can you identify the different sections of the file?
What about Quarto?
Quarto is a new implementation of this same concept. It’s billed as a “next generation version of R Markdown” because it incorporates a lot of lessons learned from its development and adds other features. It also doesn’t need R to run, so it’s more language agnostic.
There is a growing number of journal templates for Quarto –both by the quarto team and by third parties– but the list is still not as comprehensive as the large collection offered by the rticles package.
Many of the features and syntax shown here work with little or no changes in Quarto.
File structure
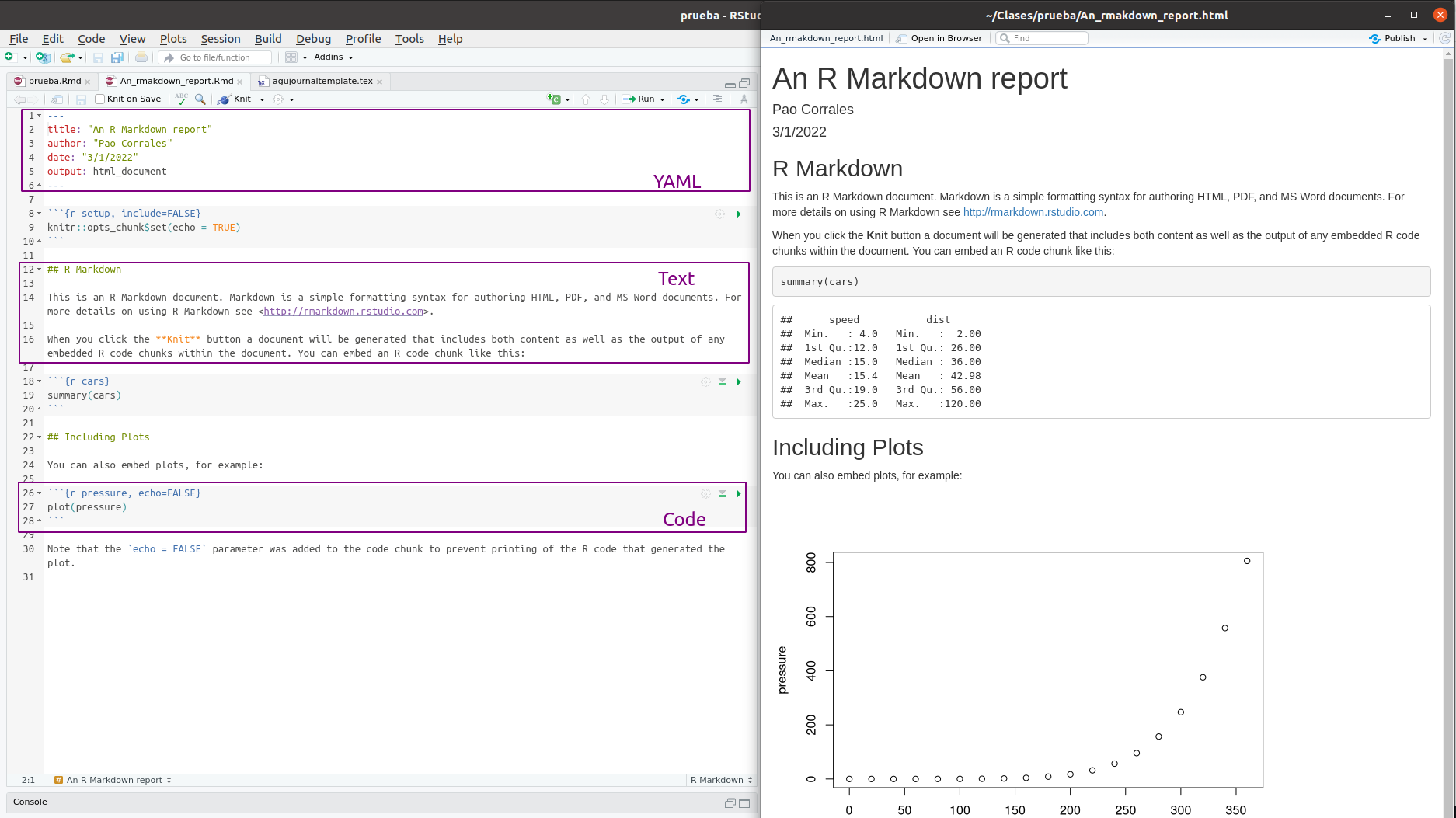
Any R Markdown file will have 3 sections or areas:
-
The top part is called the header and uses YAML syntax and includes the title and the output type (which in this case is an HTML document) and other metadata.
-
Below the header goes the document proper, which has white and grey sections. These are the two main sections that make up an R Markdown file:
-
Grey sections are code chunks. They will be evaluated in order and any output will be inserted in the output document.
-
White sections are text sections and support markdown for styling.
-
Header
The header is a series of instructions organized between three dashes (---) that define the global properties of the document, such as the title, the output format, authorship information, etc.
You can also change options associated with the output format, such as the style of the table of contents or index.
The header will grow much much larger as you start using templates and customised reports.
The YAML format allows you to define hierarchical lists in a humanly readable way. For example:
---
title: "My first markdown"
output:
html_document:
code_download: true
toc: true
toc_float: false
---
Mind the indentation!
It is very important to maintain the indentation of the elements, since it defines the hierarchy of each element. Many of the errors you’ll find when knitting will be rooted in indentation issues.
Code chunks
The R code is written inside code “chunks”.
Code chunks start with ```{r label} (where “label” is an optional and unique name) and end with ```.
Everything you include between these delimiters will be interpreted by R as code and will be executed it when the file is knitted.
Any output (graphics, tables, text, etc..) will be inserted into the final document in the same order as they are in the R Markdown file (or “roughly in the same order” if you’re knitting to PDF).
You can create a new chunk with:
- The “+C” green bottom on the top right of the document
- A very handy shortcut:
Ctrl+Alt+IorCommand+I - Writing ```{r} by hand (but why would you?)
While the code will run line by line when you knit the file, during writing it is very convenient to run individual chunks interactively as if it were in the console.
To run the line where your cursor is use the shortcut:
Ctrl+Enter or Command+Return
But you can also run the code of the whole chunk with:
Ctrl+Shift+Enter or Shift+Command+Return
By default, the output will appear immediately below the chunk.
Text
The plain text sections of an R Markdown files is interpreted as Markdown. You can get a guide to rmarkdown in this cheat sheet, but here is a minimum syntax to get you started:
- headers start with
#or##and so on (it’s important to put a space after the last#). - bold text is surrounded with
**or__. - and italic text, with
*or_.
You can also add equations and other symbols with LaTeX in line (`$E = mc^2$` looks like \(E = mc^2\)) or in its own line as:
$$
y = \mu + \sum_{i=1}^p \beta_i x_i + \epsilon
$$
It looks like:
$$ y = \mu + \sum_{i=1}^p \beta_i x_i + \epsilon $$
In-line code
You may find yourself mentioning results in the text, for example something like “the average minimum temperature for the month of March was 18 degrees”. And it is also possible that this value will change if you use a different database or if you then generate the same report for different months. The chances of you forgetting to update that “18” are high, that’s why R Markdown also has the possibility to incorporate in-line code.
Let’s imagine that you have a variable temperature to which you assign the value “18”:
temperature <- 18
To use that value in the text you have to put the name of that variable between two “`” and to indicate that it is R code this way `r temperature`.
Then if at any time the value of the variable changes, the next time you knit the document it will be updated in the text.
Chunk control
You may have noticed that code chunks in the template document include some information between the {r }.
Besides the chunk name you can also add options to control how the chuck will behave when the file is knitted.
Usually the code chunk will be included in the output, which is fine when you or the person that will read the report wants to see the code that generates those results, but it might not be what the final audience of the report might need.
It’s up to you to decide if you want to show the code or not.
To change the options of a code chunk, all you have to do is list the options inside the square brackets. For example:
```{r chunk-name, echo = FALSE, message = FALSE}
```
A particularly important set of options are the ones that control whether the code is executed and whether the result of the code will remain in the report or not:
-
echo = FALSEruns the code chunk and displays the results, but hides the code in the report. This is useful for writing reports for people who do not need to see the R code that generated the graph or table. -
include = FALSEruns the code but hides both the code and any output. It is useful to use in general configuration chunks where you load libraries. -
eval = FALSEprevents the code chunk from being run at all. Naturally, it will not display results either. It is useful for displaying example code if you are writing, for example, a document to teach R or if you want to suppress specific parts of your code.
If you are writing a report where you don’t want any code to be shown, adding echo = FALSE to each new chunk becomes tedious.
The solution is to change the option globally so that it applies to all chunks.
This is done by the knitr::opts_chunk$set() function, which sets the global options of the chunks that follow it.
You’ll find this function on the first “setup” chunk.
```{r setup, include = FALSE}
knitr::opts_chunk$set(echo = FALSE,
message = FALSE,
warning = FALSE)
```
Figures control
If the output of the chunk is a figure you have a ton of options to control its aspect. From it size, resolution, caption, etc..
You can start typing “fig.” inside the { } to explore the options.
RStudio Visual editor
Recent versions of RStudio include a Visual Editor which allows you to write Markdown markup, add citations, tables, etc. with a friendly GUI.
Managing references
If you are here to use rmarkdown to write papers or reports, you may need to add references to other papers or sources. Similarly to LaTeX, you’ll need a .bib file with all the references you want to include in the document. While we don’t have time to explore tools to create and manage this type of file, we recommend you to explore Zotero.
Provided you have the references.bib file in your project (you can try it with
this example), you need to include that in the yaml of the file:
---
title: "My first markdown"
output:
pdf_document
bibliography: references.bib
---
Each reference will have a key that defines it.
To insert a reference with, use @key, or [@key] or [@key; @key2] to insert them inside parenthesis.
It is also possible to use the LaTeX syntax but we prefer the markdown syntax to be able to knit the report to many type of outputs.
R Markdown Templates
Using rticles
If you have to write a report or document for your institution or maybe a paper for a scientific journal, you may want to use a template to change the look of the final document. Depending on the output format, templates will be different. In this section will focus on PDF files.
For scientific journals you may find the rticles package very useful. This package includes several templates –most of them contributed by the R community– so you can create journal-specific PDFs directly from RMarkdown. We recommend installing the development version from GitHub, which often includes new or updated article formats.
After installing rticles you’ll need to restart RStudio for its templates to be available in the UI.
To use rticles from RStudio, you can access the templates through File -> New File -> R Markdown.
This will open the dialogue box where you can select from one of the available templates:
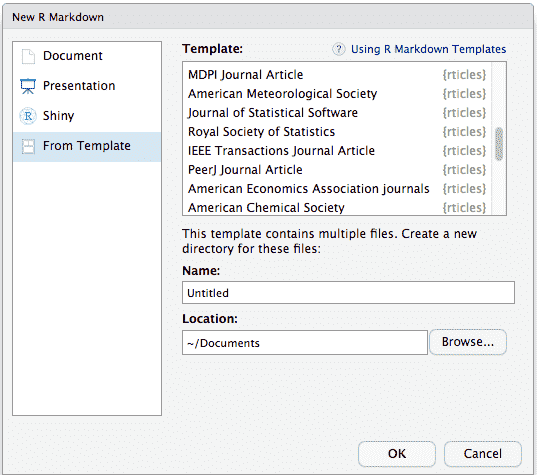
This will create a folder containing a Rmd file using the corresponding output format and all the assets required by it.
Your turn!
- Create a new R Markdown file using one template of your choosing
- Check the options in the YAML and change a few of the fields. It doesn’t have to be real information!
- Knit the document to see the output.
Beyond the usual templates
Now, what happens if rticles doesn’t have the template you need? Usually, journals provide LaTeX templates that you can use and adapt to use within R Markdown. This will require some knowledge of LaTeX and Pandoc templates, and some patience and coffee/tea/mate to deal with knitr errors. If you go through the trouble of adapting a template, consider contributing it to rticles so that others can benefit from your sweat and tears.
One estimate put the amount of work required to format and re-format academic submissions to comply with journal templates at around 230 million USD in 2001. Also, it’s not always mandatory to submit your paper using the journal template.
Adapting a template
-
Create a new R Markdown file using PDF as a output format and save it into your project (for example,
paper.Rmd). -
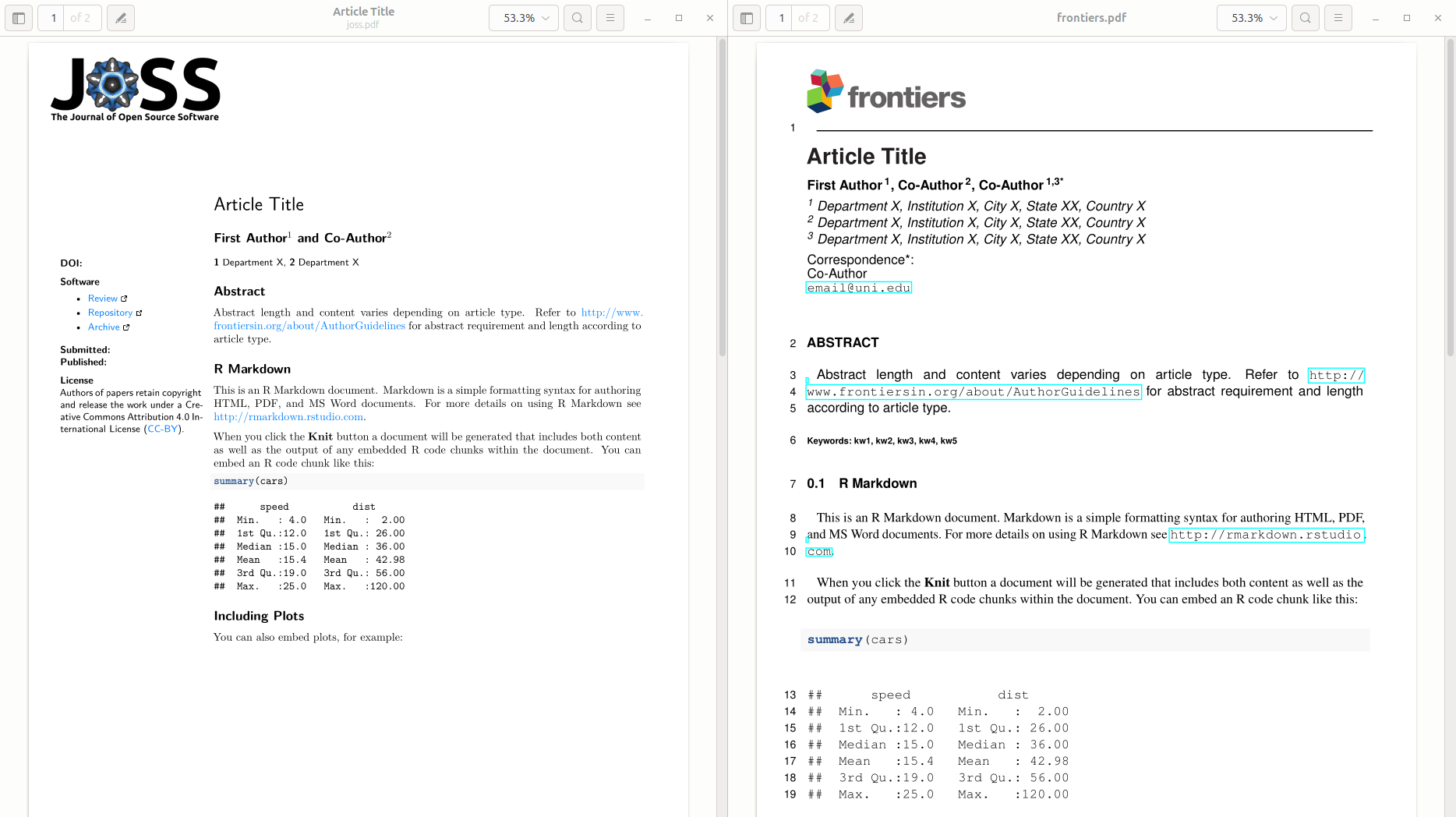
Download the AGU Geophysical Research Letters template from here. Inside the file there’s a folder with a bunch of files. Extract all those files into the same folder you saved the R Markdown. You need to have a folder in your project with these files:
. ├── agujournal2019.cls ├── agujournaltemplate.pdf ├── agujournaltemplate.synctex.gz ├── agujournaltemplate.tex ├── agutexSI2019.cls ├── paper.Rmd ├── paper.Rproj ├── si_template_2019.tex ├── trackchanges-0.7.0 └── trackchanges.sty -
Change the YAML of the R Markdown file to include the following:
--- title: "A very nice Title" author: "Pao Corrales" date: "3/1/2022" output: pdf_document: template: "agujournaltemplate.tex" pandoc_args: - "--wrap=none" ---Take care to respect the identation and that each parameter is in its own line. This new line will tell to knit to use the
agujournaltemplate.texfile as template. The template needs some tweaking, though. -
Set the global option
echo = FALSETo do this, look for the line
knitr::opts_chunk$set(echo = TRUE)and change it toknitr::opts_chunk$set(echo = FALSE). This is because the latex template doesn’t yet define the necessary environment to display code blocks, which is generally not necessary in journal articles. If you need to show code blocks, you can add`$highlighting-macros$`(see this StackOverflow answer). -
In the
agujournaltemplate.texfile, add\usepackage{hyperref}on line 63 (or anywhere before\begin{document}. Also, remove remove everything from line 174 (around% BODY TEXTto line 420 (right before\end{document}). -
Knit!
Open paper.pdf and you will see a rendered PDF in the style of the AGU journal. However, none of the contect in the R Markdown file it reflected in the PDF! You have to modify the template to connect the dots.
-
Again in the
agujournaltemplate.texfile, change\title{=enter title here=}(line 78) for\title{$title$} -
Knit again!
Now, the title in the R Markdown file and the pdf are the same.
`$title$`(without the backtics) in the LaTeX template will be replaced by the value oftitlesupplied by the YAML header. -
Let’s also include the content of the R Markdown file. In the LaTeX template, go to right before
\end{docunent}(around line 174) and add`$body$`(without the backticks). The special variablebodywill be replaced by the actual rendered contents of the R Markdown file. -
Knit again!
This time you’ll see the sample R Markdown document in the PDF.
-
You can add new options to the YAML header and then use the same trick so they are used by the LaTeX template. For example, add a new option called “abstract” and write some text, like this:
--- title: "A very nice Title" author: "Pao Corrales" date: "3/1/2022" output: pdf_document: template: "agujournaltemplate.tex" pandoc_args: - "--wrap=none" abstract: "This is the very interesting abstract." ---Then, in the LaTeX template search for
[ enter your Abstract here ](~line 166) and replace it with`$abstract$`(without backticks). -
One final knit to see the result!
-
(Optional) Add more variables, such as a Plain Language Summary, author, mail of the corresponding author.
Having problems? You can watch a video on how to solve this activity here and follow along (the video was made with an slightly older version of the template but everything is almost the same except for the precise line numbers).
Pandoc template syntax
The template uses something called Pandoc Templates, which has a specific syntax. You can read more in the documentation.